How can we quantify the attribution of antibiotic resistance to detrimental patient outcome? Some believe that in 2050, 10 million people will die every year because of antibiotic resistance!
But is antibiotic resistance always the cause of death? And if so, how can we explain detrimental outcomes of infections treated with antibiotics, for which the causative pathogen is considered susceptible?
I remember a 53-year-old patient battling aggressive leukaemia. Despite two attempts at stem cell transplantation, the patient showed no response and remained deeply neutropenic for months. The patient eventually succumbed to an uncontrollable osteomyelitis of the skull caused by pan-resistant Pseudomonas aeruginosa. The bacterium had gradually acquired resistance to all antibiotic classes that were available and used. It was sad, but even with some new antibiotics, I doubt we could have changed the outcome.
So, what factors determines the outcome of an infection? And to what extent does antibiotic resistance contribute?
Given the nature of the questions, a randomised approach will not give us answers, leading us to rely on observational data, despite its inherent bias potential. Only through the meticulous adjustment of all variables that may contribute to the outcome (such as the capabilities of the surgeon performing source control) can we potentially determine the role of antibiotic resistance in a patient’s death.
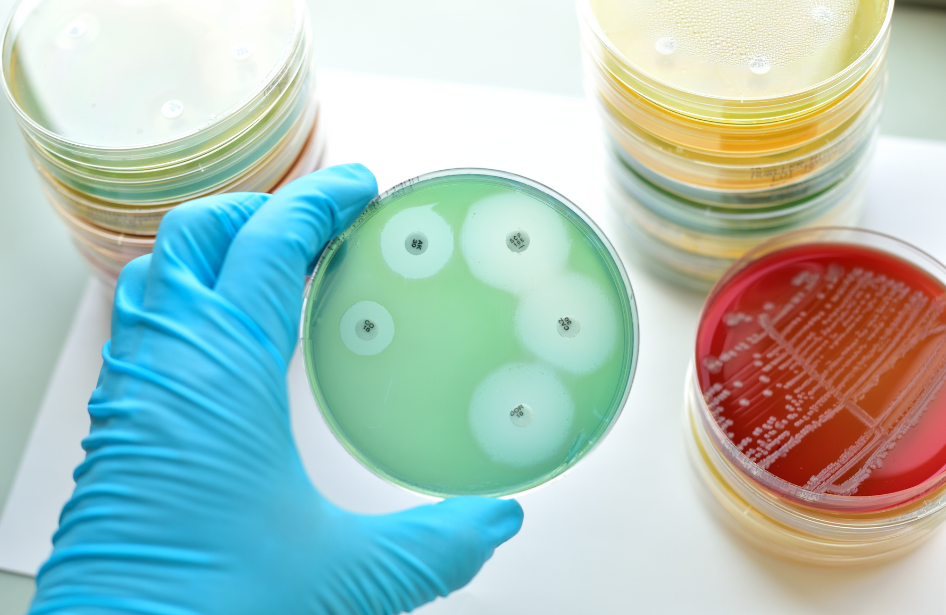
To illustrate the complexity of this issue, Wouter Rottier, a former PhD student in our group, introduced me – years ago – to the Direct Acyclic Graph (DAG). According to Wikipedia: “In mathematics, particularly graph theory, and computer science, a directed acyclic graph is a directed graph with no directed cycles. That is, it consists of vertices and edges, with each edge directed from one vertex to another, such that following those directions will never form a closed loop.”
The DAG proved quite useful in explaining that antibiotic resistance is not the only factor that determines patient outcomes. Rather, it highlighted that the interaction among various factors can be rather complex. I have presented the following slide at numerous presentations – it is confusingly convincing, don’t you think?
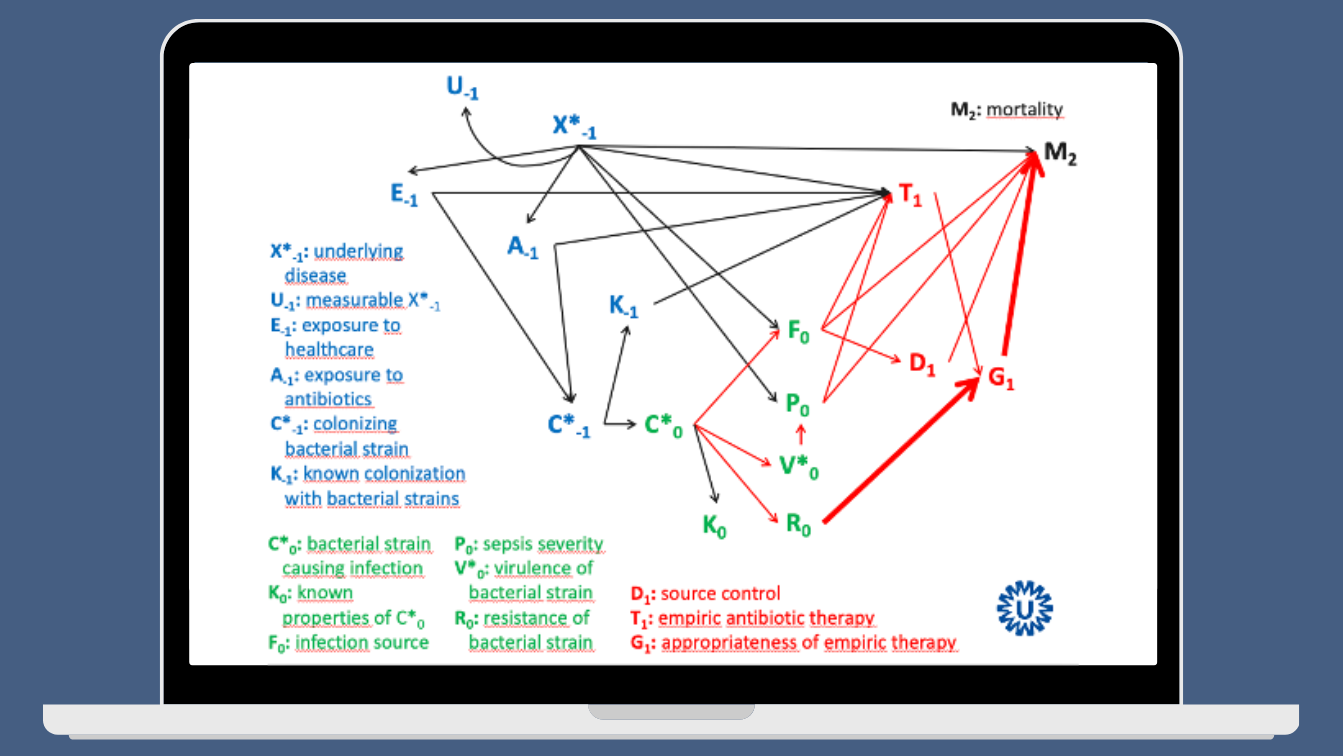
This month’s Clinical Microbiology and Infection published a Delphi survey, based on a systematic review, that defined a minimum set of risk factors that should be assessed and reported in all studies assessing survival in bacteraemia. The systematic review included 62 studies, comprising more than 300,000 patients, from which a list of 17 risk factors was derived. These factors showed statistically significant associations with all-cause mortality in most studies, and a DAG was created to illustrate this:
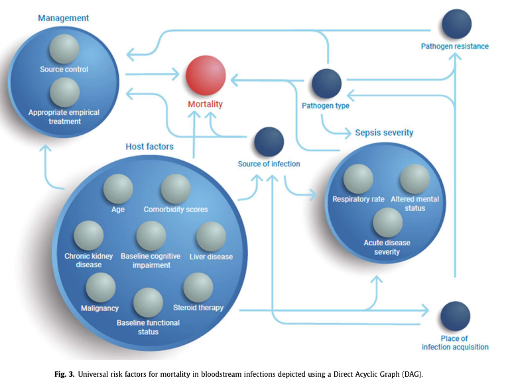
This looks much better compared to my old DAG, but in essence it shares similar characteristics. “Appropriate empirical treatment” stands out among the 17 factors identified. As one of the experts on the Delphi panel, I echo the authors’ recommendation to consistently report on these variables when evaluating mortality in patients with bacteraemia or sepsis, and to always adjust for these factors when assessing “interventions” in observational studies.
Interestingly, consultation by an infectious disease specialist in patients with S. aureus bacteraemia was absent from the DAG, as the expert panel deemed it ‘clearly not a causal risk factor’.
While antibiotic resistance will always pose a threat to the successful infection treatment, numerous other variables can influence patient outcomes from infection, with some of them amendable for interventions. The DAG is a valuable tool that helps us better understand complexity and improve the way we evaluate data from observational studies.